Three new scientists join our group
Three new post-docs, Daniel, Jeremy, and Koyo joined our team. Check their bios on our lab members page!

Nick Gilbert's Lab, Edinburgh
In mammalian cells DNA is packaged into chromatin. In our lab we study DNA and chromatin structure to understand gene regulation and genome stability.
Hear a podcast and read a transcript from The Naked Scientists, presented by Nick Gilbert - Twisting DNA.
View a video from The University of Edinburgh about our Chromatin Biology research, presented by Nick Gilbert - Packaging The Genome.
Read more about our research.
Three new scientists join our group
Three new post-docs, Daniel, Jeremy, and Koyo joined our team. Check their bios on our lab members page!

Charles joins the team
Charles is a new postdoc joining us fromt TestED, the university COVID testing scheme, he will be studying T independent response to the SARS-COV2 membrane protein. Check their bio on our lab members page.

Rafal joins the team
Rafal is a new postdoc - he will be investigating transcriptional noise and the role of SAF-A on influencing chromatin structure as well as determining how mutations in SAF-A affect its role. Check their bio on our lab members page.

Two new scientists join our team
September has been busy for @Chromatinlab with two new scientists joining our team. Tas has started a PhD, funded by the MRC Precision Medicine programme, and will investigate how chromatin structure is important for centromere formation. James is a new postdoc - he's bringing his expertise in molecular biology and 3C techniques to understand the 3D structure of gene promoters. Check their bios on our lab members page.
Chromatin meets Physics: New Wellcome grant for the lab
In this new 5 year study we will investigate how chromatin structure affects transcription.
A direct link between structure and transcription has been postulated to exist on the basis of long-standing experiments, but it has never been demonstrated in practice, as laboratory experiments on their own lack the sufficient resolution. The key novelty in our approach is the synergistic combination of our experimental and modelling techniques, which allows us to answer questions which it was previously impossible to address. For instance, we are now using simulations to predict the 3D structure and transcriptional activity of all human genes in a given cell type, validating the results experimentally as we go. As our project develops we hope to develop a new understanding of how mutations in chromatin-associated proteins, regulatory factors or DNA elements could lead to genetic diseases and cancer.
Jon joins lab
Jon Stocks has joined our group as a PhD student to work on the relationship between RNA and chromatin. Exciting time for this study, lots of new ideas and different ways to think about chromatin organisation.
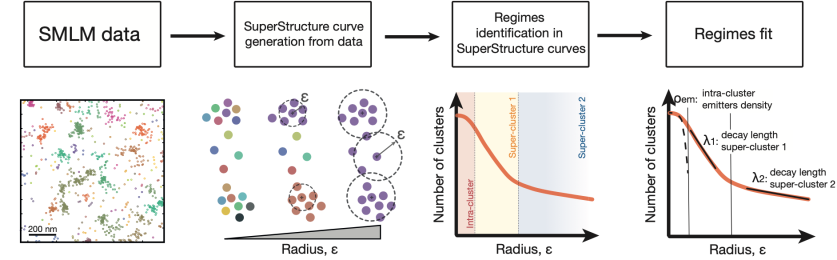
SuperStructure - new paper in Journal of Cell Biology
Read the full paper: Parameter-free molecular super-structures quantification in single-molecule localization microscopy.
In this new work, we introduce SuperStructure, a parameter-free algorithm to analyse complex biological data from super-resolution microscopy and extract patterns beyond-simple-clustering! In particular then, we suggest that mesoscopic structures formed by interconnected protein clusters are common within the nucleus and have an important role in the organisation and function of the genome.
This is an interdisciplinary and collaborative work by Mattia, Elena and Nick from the our lab, Davide Michieletto from the Department of Physics and Astronomy and Sebastian van de Linde from the University of Strathclyde.

Collaboration with Yanick Crow's group published in Nature Genetics
Read the full paper: cGAS-mediated induction of type I interferon due to inborn errors of histone pre-mRNA processing.
In this paper we show that mutations in two genes involved in histone protein expression cause Aicardi-Goutieres syndrome. Our experiments suggest that altered levels of linker histones affect the structure of chromatin that is then recognised by cGAS triggering a type 1 interferon response.
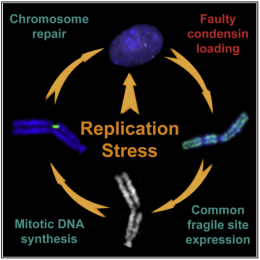
Fragile sites have a link to condensin
Read Lora's Paper: Common Fragile Sites Are Characterized by Faulty Condensin Loading after Replication Stress.
Common fragiles are regions of the genome that show an aberrant chromatin structure after replication stress. We didn't know the underlying mechanism but Lora has now shown that condensin is not properly recruited to these regions, causing the chromosome to not fold correctly. This isn't the result we were expecting and shows there are still many things to learn about chromosome structure.
New clinical PhD student
Patrick joins the lab as a Edinburgh Clinical Academic Track training fellow to do a PhD. Patrick will examine how Epstein-Barr virus sits latent in the nucleus of our cells, and will help to complete a study examining coronavirus antibodies.
Important perspective on mechanistic modeling of chromatin
Read the full article: Mechanistic modeling of chromatin folding to understand function
Over the last few years we have been working closely with Davide Marenduzzo's group in physics to understand chromatin folding. However, working at the interface between fields is challenging so we have published a detailed perspective on mechanistic modeling of chromatin. We have written the article to be understandable by biologists so they can interpret the physics methods we use to model the structure of chromatin. As we begin to ask more detailed questions we will have to rely more on modeling - particularly in a field like chromatin. But understanding each other's language is crucial.
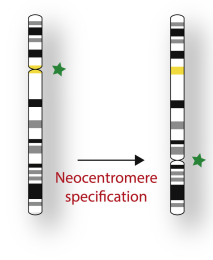
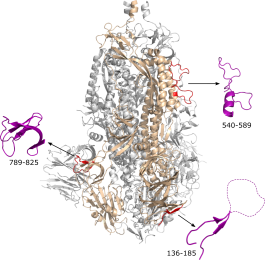
Centromere chromatin structure – Lessons from neocentromeres
Read Catherine's review: Centromere chromatin structure – Lessons from neocentromeres.
Centromeres are highly specialized genomic loci that function during mitosis to maintain genome stability. Formed primarily on repetitive α-satellite DNA sequence characterisation of native centromeric chromatin structure has remained challenging. Fortuitously, neocentromeres are formed on a unique DNA sequence and represent an excellent model to interrogate centromeric chromatin structure. This review uncovers the specific findings from independent neocentromere studies that have advanced our understanding of canonical centromere chromatin structure.

Negative supercoil at gene boundaries modulates gene topology
Read the full paper: Negative supercoil at gene boundaries modulates gene topology.
Exciting collaboration with Yathish Achar and Marco Foiani in Italy, studying how DNA supercoiling regulates the structure of genes.

4D Epigenome Conference, Venice 2019
Nick, Davide, Mattia and Elaine attended the 4D epigenome conference in Venice to discuss the principles that control the dynamic organisation of the genome. Nick presented work on the organisation of chromatin by SAF-A and RNA. Davide discussed the physical principles of retroviral DNA integration and its use as a probe for chromatin structure. Mattia presented a poster on design principles of chromosomes. Elaine presented a poster and a short talk on the impacts of chromatin topology on large-scale chromatin structures.
Thank you to Davide and the rest of the organisers for putting together such an interesting programme of talks and inspiring discussion on the complex questions at the interface between physics and biology.

DNA Packaging: Nucleosome and Chromatin
Read Nick and Jim's editorial free and find the rest of the articles: DNA Packaging: Nucleosome and Chromatin.
The latest issue of Essays in Biochemistry focuses on DNA packaging into chromatin. It is edited by our very own, Nick Gilbert and Jim Allan, with cover art from Shelagh Boyle.
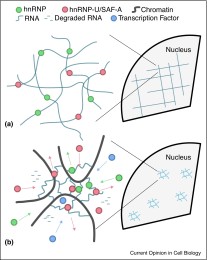
Role of nuclear RNA in regulating chromatin structure and transcription
Read the review paper: Role of nuclear RNA in regulating chromatin structure and transcription.
It is becoming clear that the 3D organisation of chromatin is a key component in genome regulation. In our latest review paper, Davide and Nick discuss recent evidence suggesting that RNA has an important role in this dynamically controlling this organisation, as well as in shaping large-scale chromatin structure.

Conference: 3D Genome - Gene Regulation and Disease
Nick attended the Keynote Symposia on Molecular and Cellular Biology for the conference 3D Genome: Gene Regulation and Disease. He presented some of the latest work from the lab: Chromatin-Associated RNA Recycling by XRN2 Regulates Transcription and Chromosome Structure.
What a beautiful location to discuss science!

Lab Retreat 2019
Here's a photo of us at our 2017 lab retreat to Auchinleck House. We're back there next week to enjoy a fun week of science - discussing our current research, new techniques and generating new ideas as a team. Hopefully the nice weather holds up for some relaxing walks nearby!
Ryu-suke's review
Read the full review: RNA: Nuclear Glue for Folding the Genome.
Ryu-suke's review on a dynamic nuclear meshwork is now published. Here, Ryu-suke reviews the role of nuclear RNAs in organising interphase chromatin structure and contrasts the historical static nuclear matrix model with the emerging dynamic nuclear mesh model.

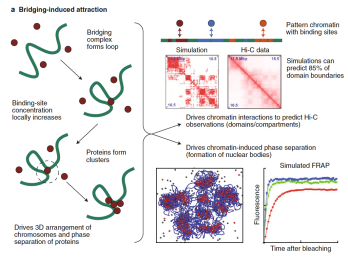
Adam's picture of HiP-HoP chromatin makes the cover of Molecular Cell
In this issue of Molecular Cell, Buckle et al. (pp.786–797) describe a “highly predictive heteromorphic polymer” (HiP-HoP) model which uses epigenetic and protein binding data to predict the 3D organization of complex gene loci. The image represents a chromatin polymer, simulated as steel beads joined together by flexible springs, within a Waddington landscape. The simulated chromatin fiber has a variable structure with regions of different stiffness generated by additional springs. Two regions of the fiber are shown being brought together to form a loop by bridging transcription factors. Cover design is by Adam Buckle.
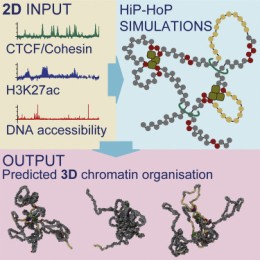
HiP-HoP: A new inter-disciplinary approach to predict complex 3D genome folding
Read the paper here: Polymer Simulations of Heteromorphic Chromatin Predict the 3D Folding of Complex Genomic Loci.
Adam and Chris have a paper in the November 2018 issue of Molecular Cell, describing a method for prediction of the 3D folding of chromatin.
Inside every cell DNA is wrapped up with proteins to form a structure called chromatin. How chromatin is folded is important for gene regulation and controls how proteins are made in different cell types. Although chromatin folding in cells is complex, chromatin fibres behave much like simple polymers. To investigate the properties important for chromatin folding we setup a collaboration with polymer physicists, to model how chromatin folds at specific genes including those important for human disease. We first collected information about how proteins bound to the DNA at these genes of interest and painted this information onto our computer based 3D polymer simulations. Then, using current knowledge of genome organisation, we added different physical properties to the polymer, such as regions with a more crumpled structure and allowed the computer simulated chromatin polymers to fold-up 100s of different times. We compared the outcome of these simulations to the physical 3D folding of chromatin inside real cells to test how well the model predicted the real 3D structure. The simulations showed us there was striking variability in the shape and folding of chromatin in individual cells, especially at active gene regions and this maybe important for regulating gene expression. We named this method the “highly predictive heteromorphic polymer model” or HiP-HoP model. HiP-HoP now allows us to accurately predict 3D folding using commonly available 2D data and understand some of the fundamental principles that organise DNA inside cells. We are now applying this method to understand how chromatin folding changes in disease, and how this in turn affects the expression of individual genes.
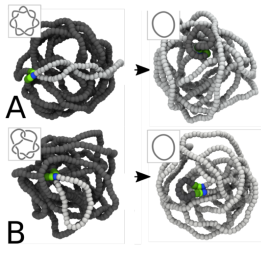
New paper from Davide
Read the paper here: Synergy of SMC and Topoisomerase Creates a Universal Pathway to Simplify Genome Topology.
SMC co-operates with TopoII to efficiently remove knots and links from in vivo chromatin.
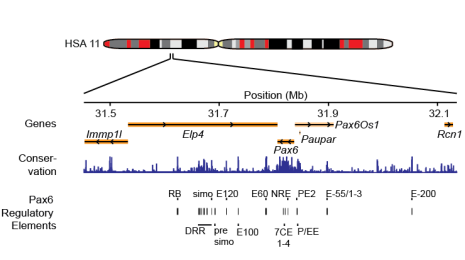
Adam's paper investigating Pax6 gene regulation
Read the paper here: Functional characteristics of novel pancreatic Pax6 regulatory elements.
Adam's paper investigating Pax6 cis-regulatory elements in human and mouse was published at Human Molecular Genetics.

Biophysics workshop
We've been enjoying the second day of the Biophysics of Epigenetics and Chromatin workshop organised by Davide Michieletto from the lab!
Nick's latest research
Read the paper here: Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation.
Nick's latest paper has been published in the Journal of Cell Biology.

Issy in 3MT University of Edinburgh final
Issy scooped 2nd place in the University of Edinburgh final of the 3 Minute Thesis Competition, impressing the judges with her 'Great Genetic Bake Off' to explain how she is researching the role of chromatin structure in meiotic recombination!
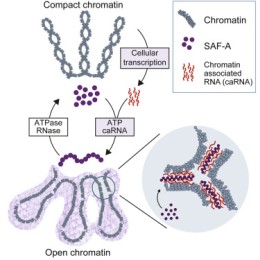
SAF-A Cell paper
Ryu-Suke's latest research on the role of SAF-A in the regulation of chromatin structure was published in Cell: SAF-A Regulates Interphase Chromosome Structure through Oligomerization with Chromatin-Associated RNAs.
Issy is 3MT winner
Congratulations to Issy for winning the IGMM's Three Minute Thesis competition with her 'Great Genetic Bake Off'! She will be going on to compete in the University of Edinburgh's College of Medicine and Veterinary Medicine round. Watch her fantastic talk here:

Nick at Gordon Research Conference
Nick talks about our latest research at the Chromatin Structure and Function, Gordon Research Conference.
Lora's review
Read Lora’s review in Kovalchuk I and Kovalchuk O (Eds), Genome Stability: Chromatin, nuclear organization and genome stability in mammals.
New PhD Students
New lab members Kate and Issy join us to do their PhDs on chromatin structure and genome stability.

Gargunnock House Retreat
Nick’s Lab join forces with Shaun Cowley’s group for our annual retreat at Gargunnock House. Check out our photos on Flickr!

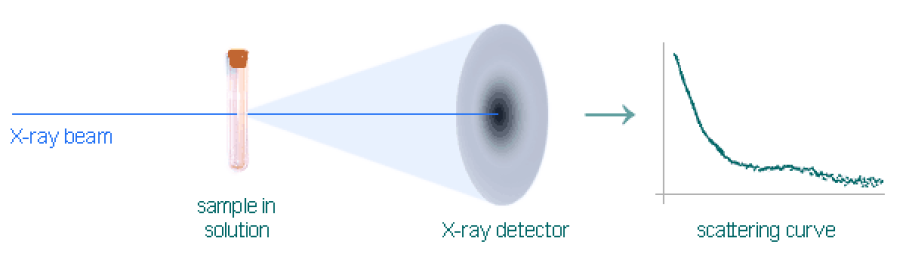
Diamond Light Source
Nick and Jim visit Diamond Light Source to investigate folding properties of chromatin fibres.
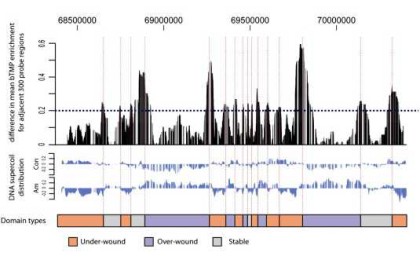
Analysing DNA supercoiling
Sam Corless discusses our bioinformatics methods for Profiling DNA Supercoiling Domains in vivo, published in Genomics Data.
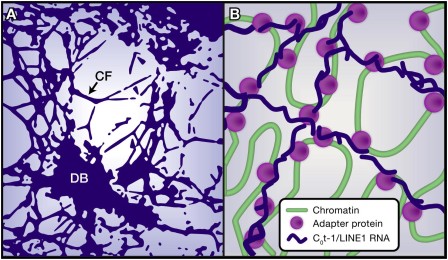
Interphase Chromatin LINEd with RNA
Interesting paper in Cell from the Lawrence lab. Read our preview Interphase Chromatin LINEd with RNA.
Supercoiling in DNA and Chromatin
Read our review Supercoiling in DNA and Chromatin in Current Opinions in Genetics & Development.
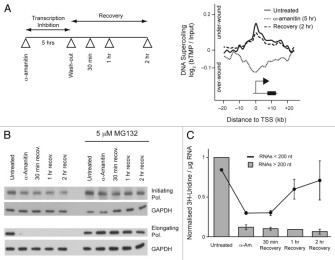
Divergent RNA transcription: A role in promoter unwinding?
Our work’s review Divergent RNA transcription: A role in promoter unwinding? was published in Transcription.
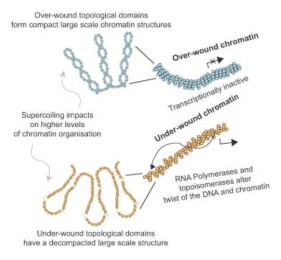
Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures
Our work Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures was published in Nature Structural & Molecular Biology.